
OmeSeq:
A Paradigm Shift In Next-Generation Sequencing
OmeSeq Technology
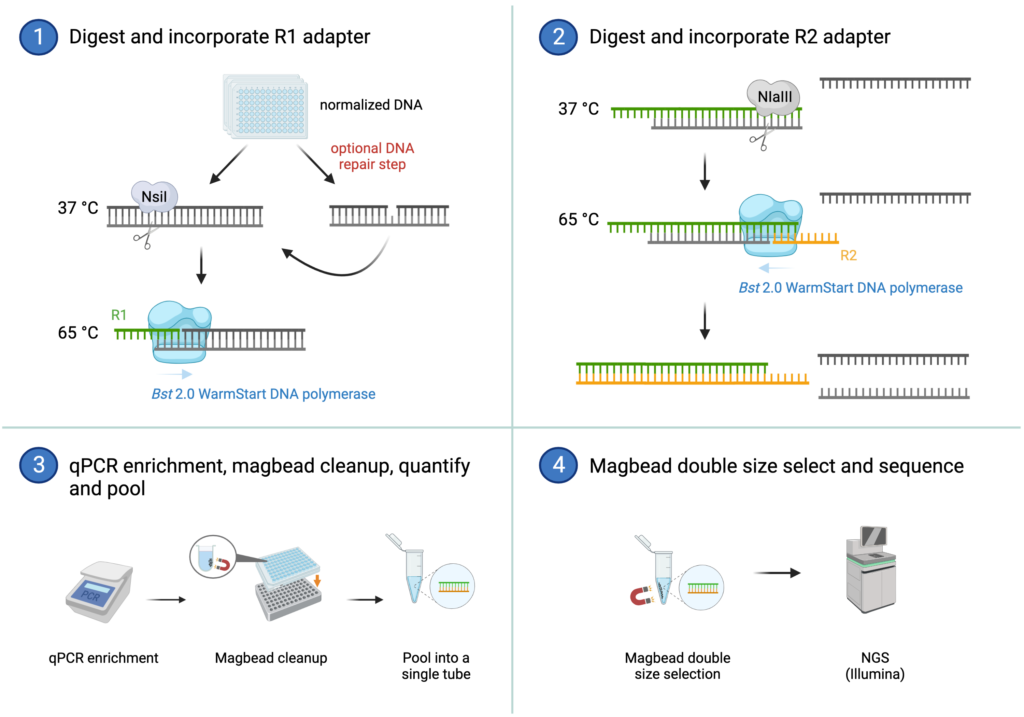
OmeSeq-qRRS (quantitative Reduced Representation Sequencing; patent pending) is a novel library preparation approach for NGS-based genotyping and metagenome profiling that mitigates current challenges and limitations of other NGS-based applications. Due to its unique adapter design and use of isothermal amplification, OmeSeq provides distinct benefits that include unparalleled read quality with high-fidelity base calls across entire read lengths. The cost-effective and quantitative assays are offered in two formats: (1) “OmeSeq-qRRS”, and (2) “OmeSeq-array” or targeted OmeSeq-qRRS (i.e., in solution oligo array). More information on each method is provided below.
OmeSeq-qRRS
Based on dual restriction enzyme digestion and isothermal amplification (ligation-free), a small fraction (scalable density) of the genome/metagenome is sequenced in gene-rich regions. Several unique features of the protocol (see below) distinguish it from other commonly used methods, such as multiplexed-pcr, GBS/ddRAD-Seq, and Capture-Seq.
OmeSeq-array
Considering a large proportion of qRRS-derived fragments/reads tend to be monomorphic, a “targeted OmeSeq-qRRS” uses inexpensive/short adapters and isothermal amplification to capture polymorphic regions. This leads to a decrease in cost of sequencing and significantly reduced computational burden. Adapter probe design/cost is very cost-effective due to a novel multiplexed-adapter synthesis method. OmeSeq-array has no limitation on SNP density; compare to other targeted sequencing approaches that are limited to a few thousand.
Key Features
Library preparation cost is $10 per sample. NGS cost is $1 to $10, depending on plex level and library type (i.e., OmeSeq-qRRS or OmeSeq-array) | |
Adapter design ensures potential chimeric reads are eliminated from library | |
Isothermal amplification and final qPCR enrichment minimizes PCR bias and permits accurate estimation of allelic ratios for dosage-sensitive assays | |
The absence of a denaturation step maintains native dsDNA structure and ensures perfect adapter hybridization/matching at high annealing temperatures (65℃) | |
Capable of multiplexing 96 – 9,216 samples. More than 9,216 is possible upon request | |
Unique adapter design generates high quality base calls across the entire length of reads | |
Degraded DNA with fragments no longer than 1-2 kb and concentrations as low as 1 ng/μl have been successfully sequenced | |
Uniform sequencing across loci and samples minimizes missing data | |
Software has been designed and implemented to complement and account for novel features of OmeSeq (see Bioinformatic Tools) |
OmeSeq Workflow
Potential Applications
Molecular breeding
-
Linkage map construction
-
Marker-assisted selection and genomic selection
-
Genetic diversity analyses
-
Phylogenetic analyses


Metagenomics profiling
-
Strain-level profiling from viruses to eukaryotes
-
Quantification of community members
-
Metagenome-enhanced association analysis and genomic prediction

Diagnostics
-
Identification of genetic disorders
-
Disease risk assessment
-
Pathogen/microbe identification
Future Applications
While current applications of OmeSeq can be applied to variant identification and metagenome profiling, the OmeSeq protocol is currently being developed for:
-
Transcriptomic profiling
-
Small RNA profiling
-
Methylome sequencing
-
NGS-based Hierarchical shotgun whole genome sequencing and assembly
(benefits of BAC-based WGS/assembly without the need for laborious BAC libraries)